Designing effective primers for quantitative Polymerase Chain Reaction (qPCR) is crucial for successful DNA or RNA quantification in various molecular biology applications. This manual provides a detailed overview of key considerations, such as primer length, melting temperature, and GC content, essential for optimal binding to target sequences. It guides users through steps like obtaining target sequences, utilizing design tools, and analyzing results to avoid common pitfalls like non-specific amplification or inappropriate amplicon sizes. The importance of testing multiple primer pairs and validating their effectiveness cannot be overstated. By following these comprehensive steps, researchers can improve the reliability of their qPCR assays significantly.
Understanding qPCR and Primer Design
 Credits: thermofisher.com
Credits: thermofisher.com
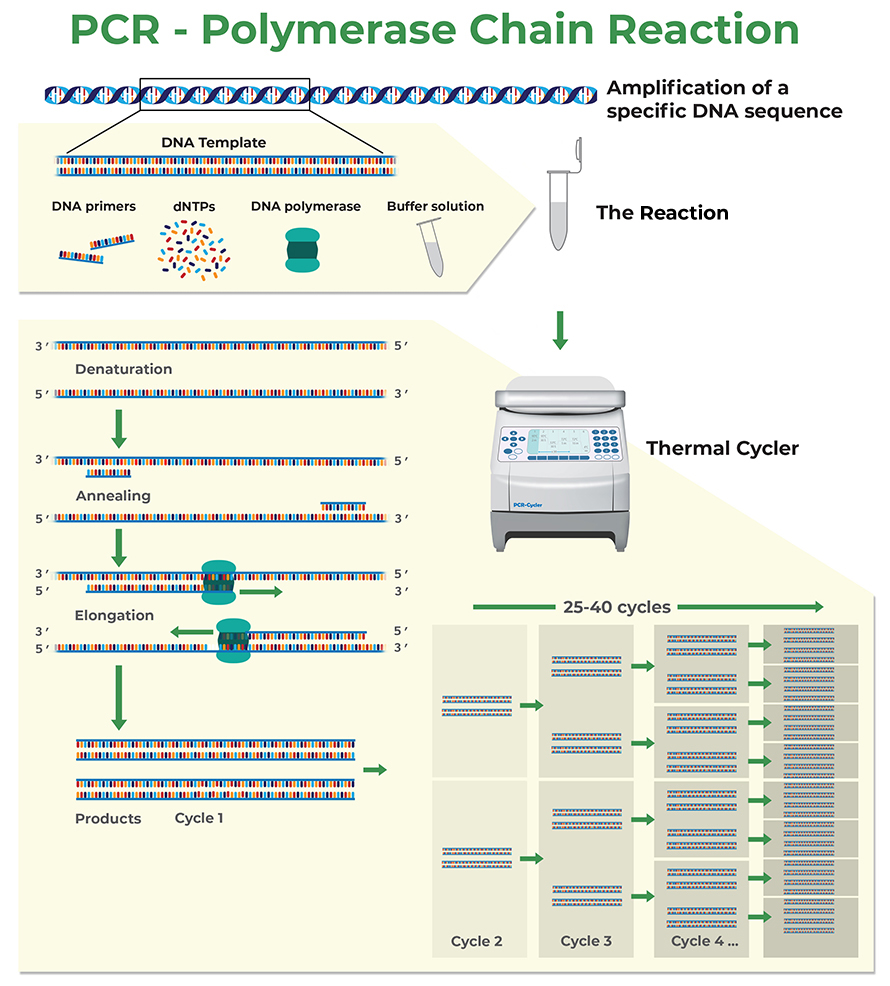
Quantitative Polymerase Chain Reaction, or qPCR, is a powerful technique in molecular biology that allows researchers to measure the quantity of specific DNA or RNA in their samples. The accuracy and reliability of qPCR results heavily depend on the design of the primers used in the amplification process. Primers are short sequences of nucleotides that bind to the target DNA or RNA, providing a starting point for the polymerase enzyme to synthesize new strands of nucleic acid.
The key to successful qPCR lies in ensuring that the designed primers are highly specific to the target sequence. Non-specific binding can lead to false positives, where the assay detects unintended sequences, ultimately skewing the results. For instance, if primers designed for a specific gene also bind to a similar sequence in a different gene, it could result in misleading quantification.
In addition to specificity, the efficiency of the amplification process is influenced by several factors, including the length and melting temperature (Tm) of the primers. A well-designed primer pair will have a balanced Tm and appropriate length to ensure they bind simultaneously during the qPCR cycle, leading to effective amplification.
Another important aspect is the analysis of the amplicon size. Generally, shorter amplicons (70 to 200 base pairs) are more efficiently amplified in qPCR, making them preferable for most applications. By understanding these fundamental principles, researchers can better design their primers to achieve accurate and reproducible results in their qPCR experiments.
Key Considerations for Primer Design
 Credits: zymoresearch.com
Credits: zymoresearch.com
When designing qPCR primers, several key factors must be kept in mind to ensure optimal performance. First, the length of the primers should be between 18 and 30 nucleotides. This range strikes a balance between specificity and efficient binding to the target sequence. For example, a primer that is too short may bind non-specifically, while one that is too long may not efficiently amplify the target.
Next, consider the melting temperature (Tm) of the primers. Ideally, the Tm should be between 60°C and 64°C, with 62°C being optimal. It’s essential that the Tm of the forward and reverse primers does not differ by more than 2°C to ensure they anneal simultaneously during the qPCR process.
GC content is another critical factor. A GC content of 40-60% is recommended as it helps maintain the stability of the primer-template complex. For instance, if the GC content is too low, the binding may not be stable, leading to poor amplification.
The size of the amplicon should also be considered. Aim for amplicons between 70 and 200 base pairs. Smaller amplicons are generally amplified more efficiently and are less prone to secondary structures that can interfere with the qPCR.
Finally, specificity is paramount. Use bioinformatics tools like Primer-BLAST to check that your primers will bind specifically to the target sequence. This step is crucial to avoid amplifying non-target sequences, which can lead to misleading results.
| Consideration | Recommendation |
|---|---|
| Length of Primers | 18-30 nucleotides |
| Melting Temperature (Tm) | 60°C – 64°C (optimal 62°C) |
| GC Content | 40-60% |
| Amplicon Size | 70-200 base pairs |
| Specificity | Use tools like Primer-BLAST |
Steps to Design qPCR Primers
 Credits: zymoresearch.com
Credits: zymoresearch.com
Start by obtaining the target sequence of the gene you want to amplify. You can find this information in databases like NCBI. Once you have the sequence, utilize primer design tools such as Primer-BLAST or Primer3. Input the nucleotide sequence into the tool, set the desired PCR product size between 70-200 bp, and specify a melting temperature (Tm) range of 60-63°C. It’s also helpful to check options that allow the primers to span exon-exon junctions, which helps prevent amplification of genomic DNA.
After generating potential primer pairs, analyze the results carefully. Ensure the 3’ end of each primer has a G or C, as this enhances binding stability. Be cautious about self-complementarity, as it can lead to unwanted primer-dimer formation.
Once you have selected suitable primer pairs, order them from a reputable supplier. It’s a good idea to order at least two sets of each pair for validation purposes. Make sure to dilute and store the primers properly before using them in your qPCR experiments.
Common Pitfalls to Avoid
One common pitfall in primer design is non-specific amplification. This occurs when primers bind to unintended sequences, leading to inaccurate results. To avoid this, always use specificity-checking tools to ensure your primers target only the desired sequence. Another issue is inappropriate amplicon size; if the primers yield amplicons that are too long, amplification efficiency may drop. Aim for amplicons between 70 and 200 base pairs for optimal results. Poor melting temperature (Tm) matching between primer pairs can also cause problems. If the Tms differ by more than 2°C, the primers may not bind simultaneously, affecting the efficiency of the reaction. Additionally, ignoring secondary structures like hairpins or dimers can interfere with amplification. Always analyze your primers for these issues before proceeding. For example, a primer pair with significant self-complementarity may form dimers, which can outcompete the target during amplification. By recognizing and addressing these pitfalls, you can improve the reliability and effectiveness of your qPCR experiments.
Additional Tips for Successful qPCR
When designing qPCR experiments, consider the use of a reference gene for normalization. This helps account for variations in sample quantity and quality, ensuring more reliable results. Choose a reference gene that is consistently expressed across all samples to improve the accuracy of your analysis. Additionally, maintaining consistent reaction conditions is crucial. Make sure to use the same reagent batch and follow the same thermal cycling parameters across all runs to minimize variability.
Another important tip is to perform a dilution series of your template DNA. This helps in assessing the efficiency of your qPCR assay and allows for the identification of any potential issues with primer binding or amplification. Ideally, your assay should have an efficiency between 90% and 110%, which can be calculated from the slope of the standard curve generated from your dilution series.
Finally, consider running all samples in duplicate or triplicate to ensure the reproducibility of your results. This practice helps in identifying outliers and improves the reliability of your data. By implementing these additional tips, you can enhance the robustness and accuracy of your qPCR experiments.
- Use high-quality reagents and enzymes.
- Optimize annealing temperatures during experiments.
- Keep reaction volumes consistent across samples.
- Design primers with adequate length (18-24 bases recommended).
- Ensure primers have a balanced GC content.
- Avoid secondary structures like hairpins in primers.
- Validate primers with control samples to ensure efficiency.
Frequently Asked Questions
1. What are qPCR primers and why are they important?
QPCR primers are short pieces of DNA that help start the copying process in quantitative PCR. They are crucial because they determine which part of the DNA you will amplify and monitor.
2. How do I choose the right sequence for my qPCR primers?
To choose the right sequence, you should consider factors like melting temperature (Tm), specificity to your target gene, and avoiding sequences that can form secondary structures.
3. What should I avoid when designing qPCR primers?
Avoid long primers, sequences with too many identical bases, regions that can form hairpins or dimers, and ensure they don’t bind to non-target regions.
4. How can I test if my designed primers will work?
You can test your primers by running a basic PCR reaction and checking if you get a single band that corresponds to your target DNA on a gel.
5. What is the optimal length for qPCR primers?
The optimal length for qPCR primers is usually between 18 to 25 nucleotides, as this length helps ensure specificity and proper binding.
TL;DR This manual provides a comprehensive guide on designing effective qPCR primers, crucial for reliable quantitative PCR results. Key considerations include primer length (18-30 nucleotides), melting temperature (Tm of 60-64°C), GC content (40-60%), and amplicon size (70-200 bp). The design process involves obtaining the target sequence, using tools like Primer-BLAST, analyzing potential primer pairs, and ordering them for testing. Common pitfalls include non-specific amplification and poor Tm matching. Additional tips for success include using probes and testing multiple primer pairs.

